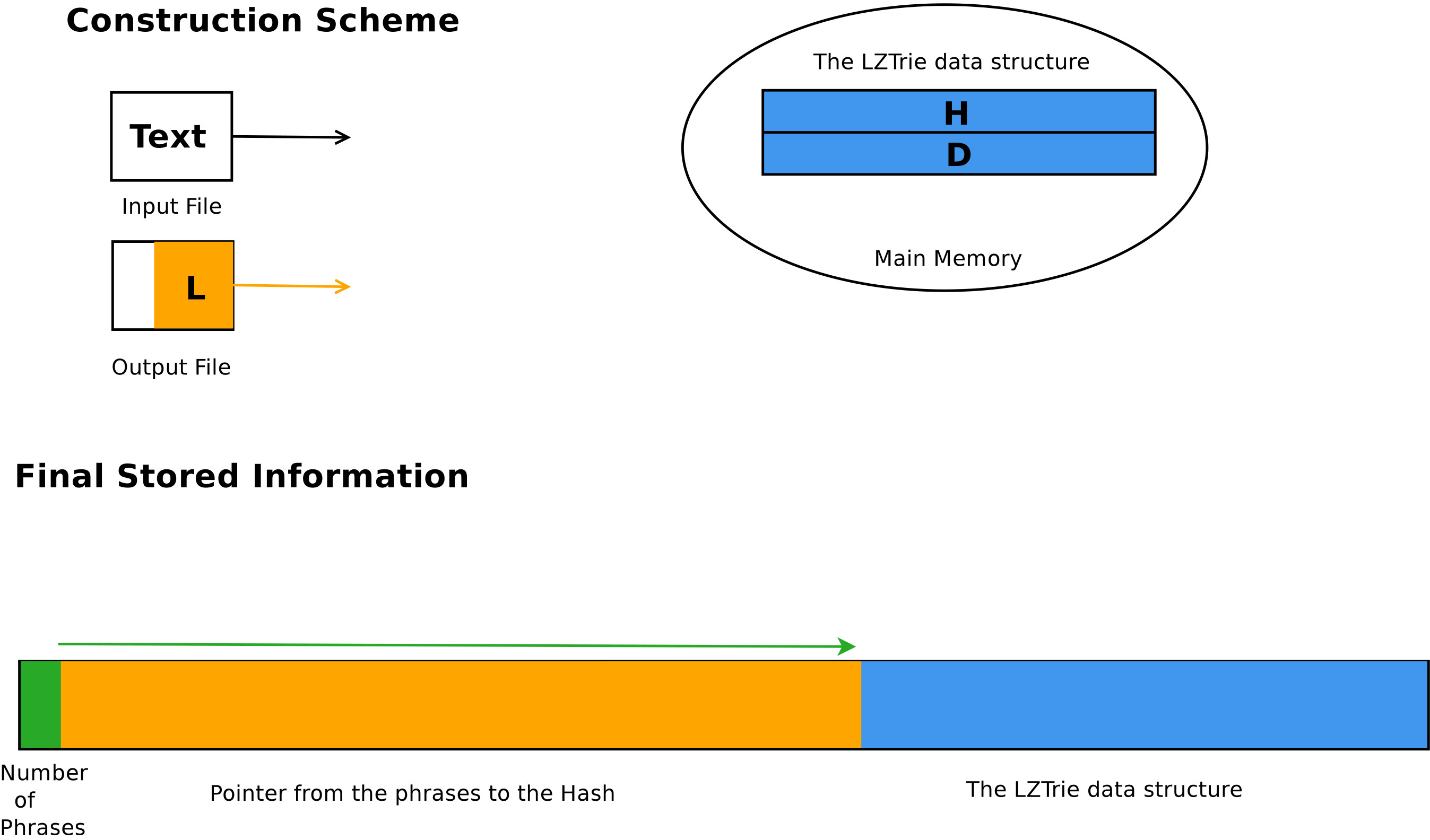
Projects
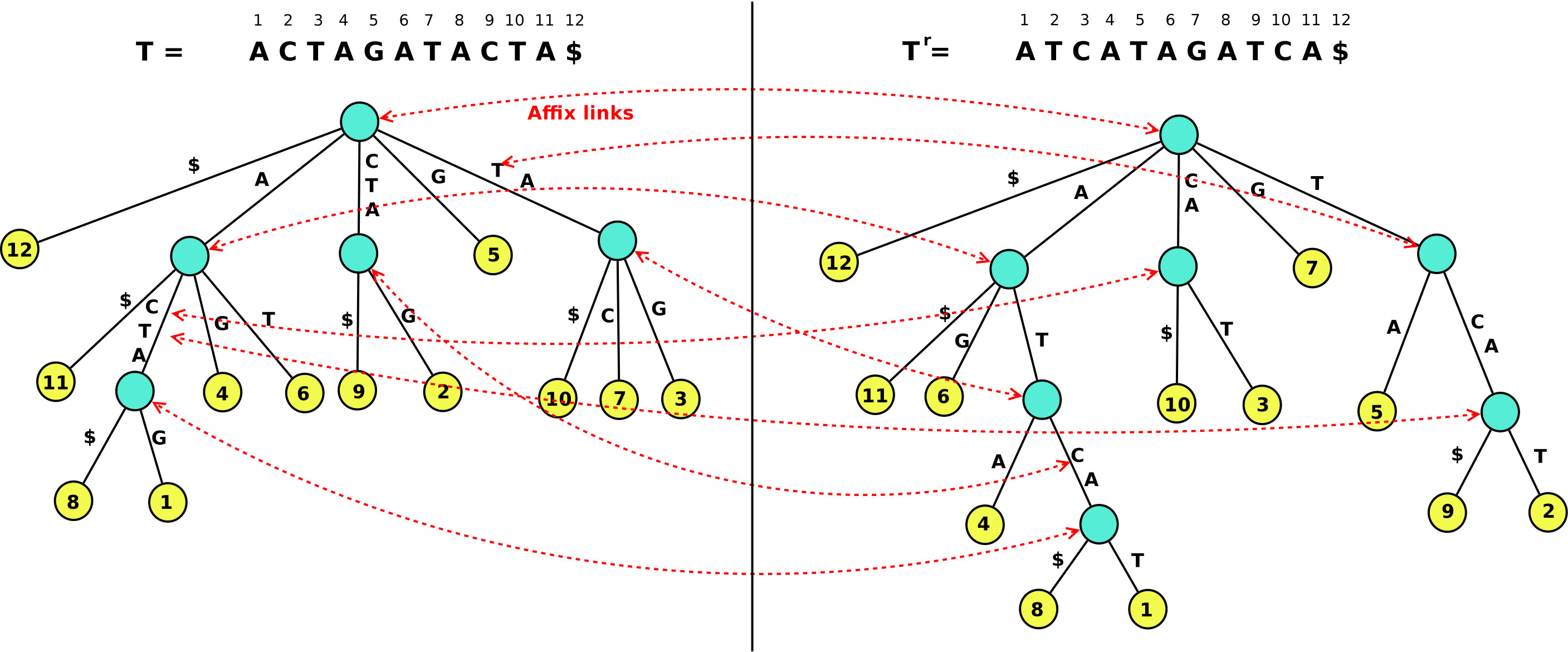
Affix Trees
Generalization of the Suffix Tree that supports full tree functionalities in both search directions
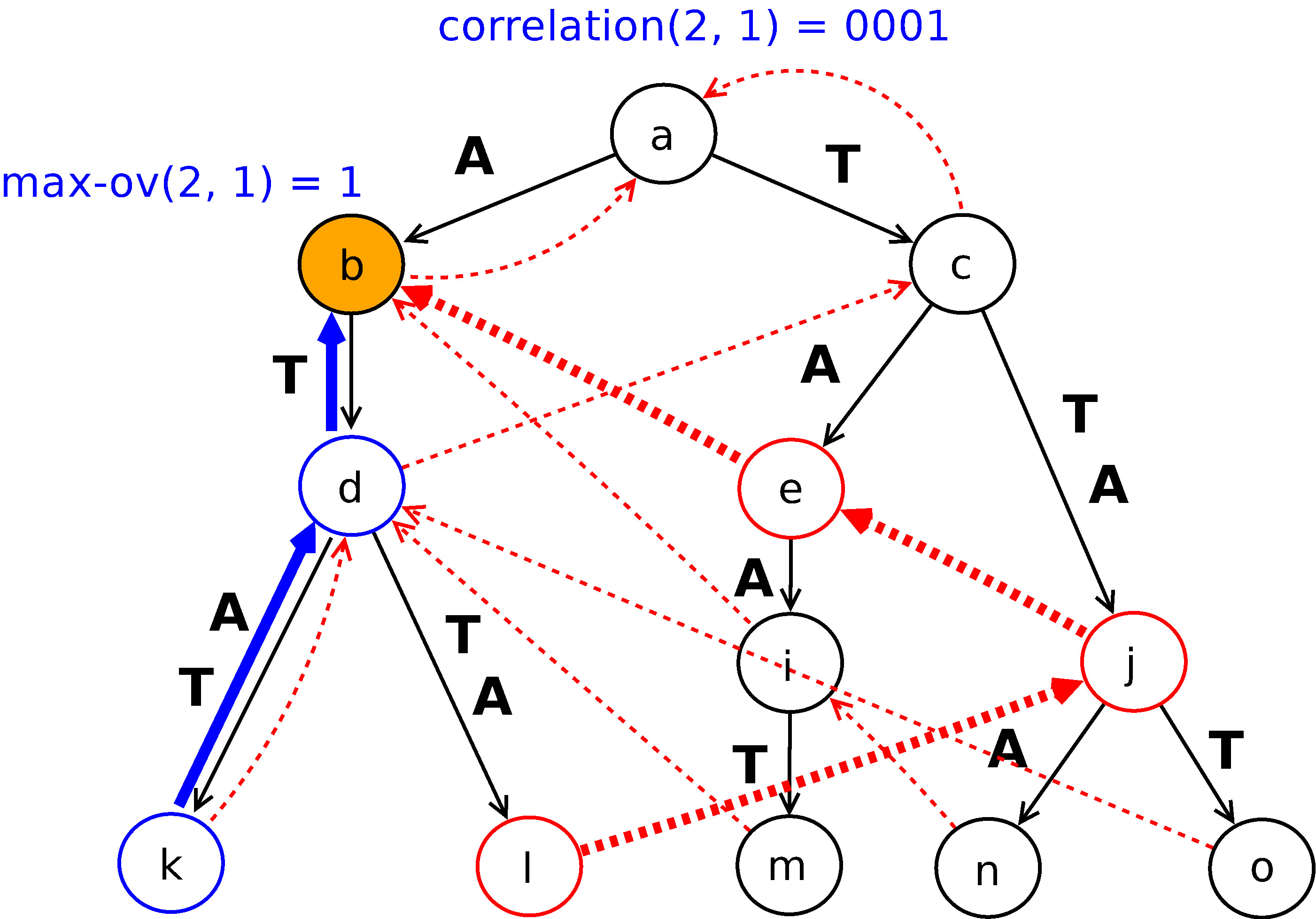
COvI
Compressed Overlap Index
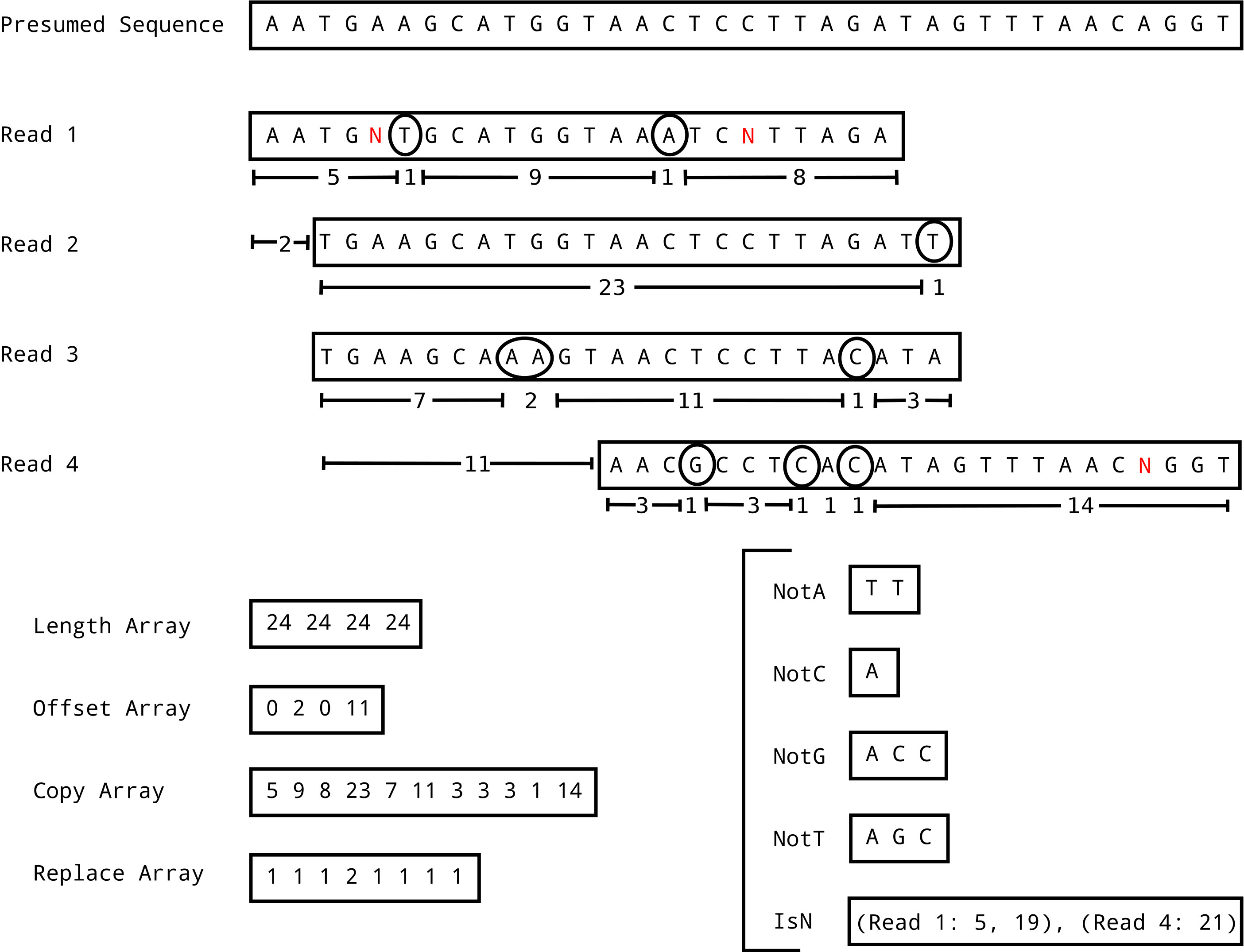
CSAM
Compressed SAM format
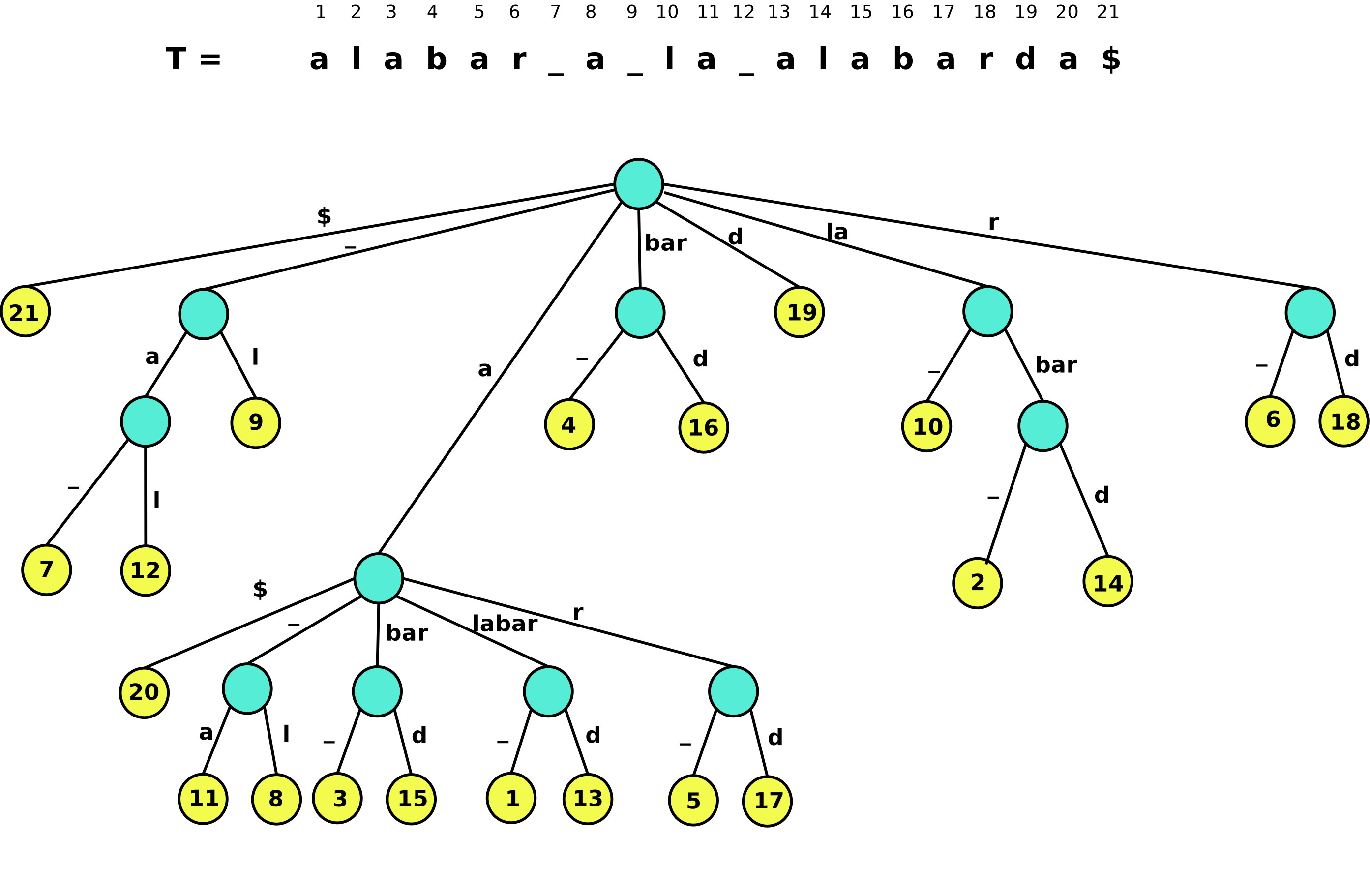
Compressed Suffix Tree
A Compressed Suffix Tree implementation
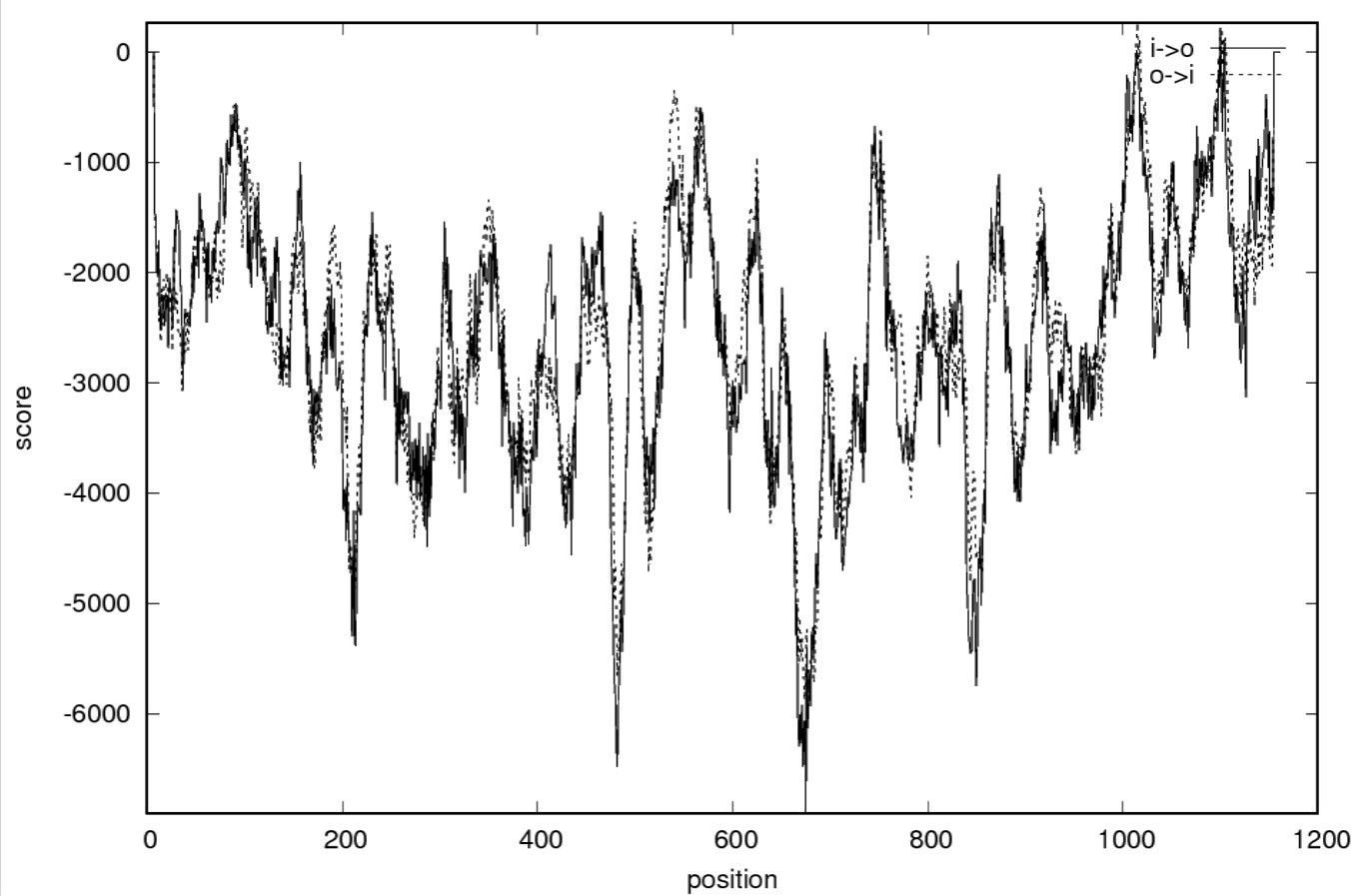
TMPred
TMPred